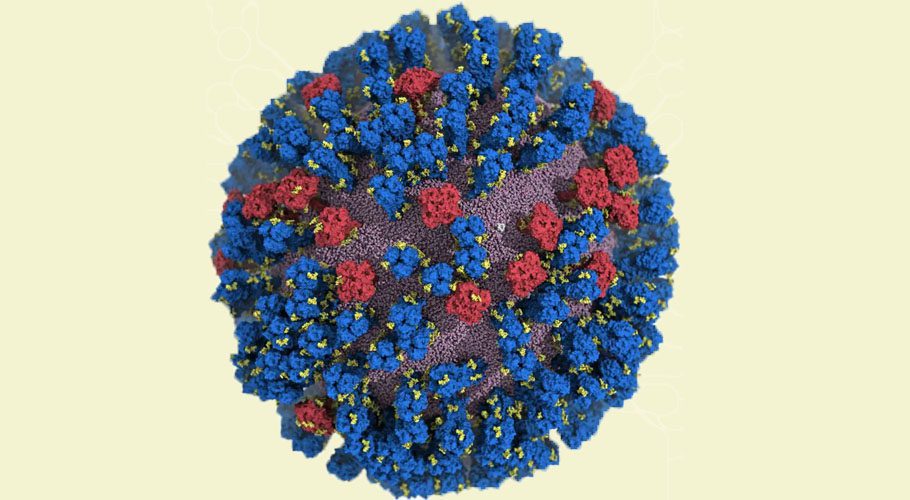
Researchers at the University of California San Diego have created an atomic-level computer model of the H1N1 virus that reveals new vulnerabilities through glycoprotein “breathing” and “tilting” movements. This work, published in ACS Central Science, suggests possible strategies for the design of future vaccines and antivirals against influenza.
Also read: Scientists discover origins of devastating childhood cancer-like disease LCH
The hemagglutinin (HA) and neuraminidase (NA) glycoproteins are the primary targets of the flu vaccine. The HA protein facilitates the virus’s attachment to host cells, while the NA protein acts as a scissor to detach the HA from the cell membrane, enabling the virus to multiply. Despite previous studies on the properties of these glycoproteins, a complete understanding of their movement does not exist.
The World Health Organization reports that there are approximately 1 billion cases of influenza annually, with 3-5 million severe cases and as many as 650,000 influenza-related respiratory fatalities worldwide. To be effective, seasonal flu vaccines must be updated each year to align with the predominant strains of the virus. When the vaccine is a match for the prevalent strain, it offers substantial protection. However, if the vaccine and virus strains are not a match, the vaccine may provide limited defense.
Also read: Treating untreatable infections: Researchers create new bacteria-killing compound
Traditionally, flu vaccines have targeted the head of the HA protein based on still images that showed the protein in a tight formation with little movement. Amaro’s model showed the dynamic nature of the HA protein and revealed a breathing movement that exposed a previously unknown site of immune response, known as an epitope.
Also read: Ultra-Processed foods linked to increased risk of cancer, death: Study
“When we first saw how dynamic these glycoproteins were, the large degree of breathing and tilting, we actually wondered if there was something wrong with our simulations,” stated Distinguished Professor of Chemistry and Biochemistry Rommie Amaro, who is the principal investigator on the project. “Once we knew our models were correct, we realized the enormous potential this discovery held. This research could be used to develop methods of keeping the protein locked open so that it would be constantly accessible to antibodies.”
Also read: Promising new method could replace injections with pills
This discovery complemented previous work from one of the paper’s co-authors, Ian A. Wilson, Hansen Professor of Structural Biology at The Scripps Research Institute, who had discovered an antibody that was broadly neutralizing — in other words, not strain-specific — and bound to a part of the protein that appeared unexposed. This suggested that the glycoproteins were more dynamic than previously thought, allowing the antibody an opportunity to attach. Simulating the breathing movement of the HA protein established a connection.
NA proteins also showed movement at the atomic level with a head-tilting movement. This provided a key insight to co-authors Julia Lederhofer and Masaru Kanekiyo at the National Institute of Allergy and Infectious Diseases. When they looked at convalescent plasma — that is, plasma from patients recovering from the flu — they found antibodies specifically targeting what is called the “dark side” of NA underneath the head. Without seeing the movement of NA proteins, it wasn’t clear how the antibodies were accessing the epitope. The simulations Amaro’s lab created showed an incredible range of motion that gave insight into how the epitope was exposed for antibody binding.
Also read: Australia approves MDMA and Psilocybin as Medicines
The H1N1 simulation Amaro’s team created contains an enormous amount of detail — 160 million atoms worth. A simulation of this size and complexity can only run on a few select machines in the world. For this work, the Amaro lab used Titan at Oak Ridge National Lab, formerly one of the largest and fastest computers in the world.
Amaro is making the data available to other researchers who can uncover even more about how the influenza virus moves, grows, and evolves. “We’re mainly interested in HA and NA, but there are other proteins, the M2 ion channel, membrane interactions, glycans, and so many other possibilities,” Amaro stated. “This also paves the way for other groups to apply similar methods to other viruses. We’ve modeled SARS-CoV-2 in the past and now H1N1, but there are other flu variants, MERS, RSV, HIV — this is just the beginning.”